|
|
|
|
|
|
|
|
|
|
|
|
|
1. Downy Mildew Genome-wide association study (GWAS) using 9,783 GBS-generated SNPs across a panel of 174 spinach genotypes led to identify loci governing resistance to downy mildew (DM) pathogen (Peronospora effusa (syn. P. farinosa f. sp. spinaciae, Pfs) race 13 on chromosome 3 in the spinach hybrids T-Bird, Swan, Squirrel, and Tonga (Bhattarai et al. 2020a) (Figure 1). GWAS was also performed in 172 spinach genotypes using 10,788 GBS-generated SNPs in a population segregating for resistance to DM Pfs race 16, derived from a cross of cvs. Whale and Lazio. SNP markers and genetic alleles were identified in the same region on chromosome 3 for Pfs 16 resistance (Bhattarai et al. 2021a). 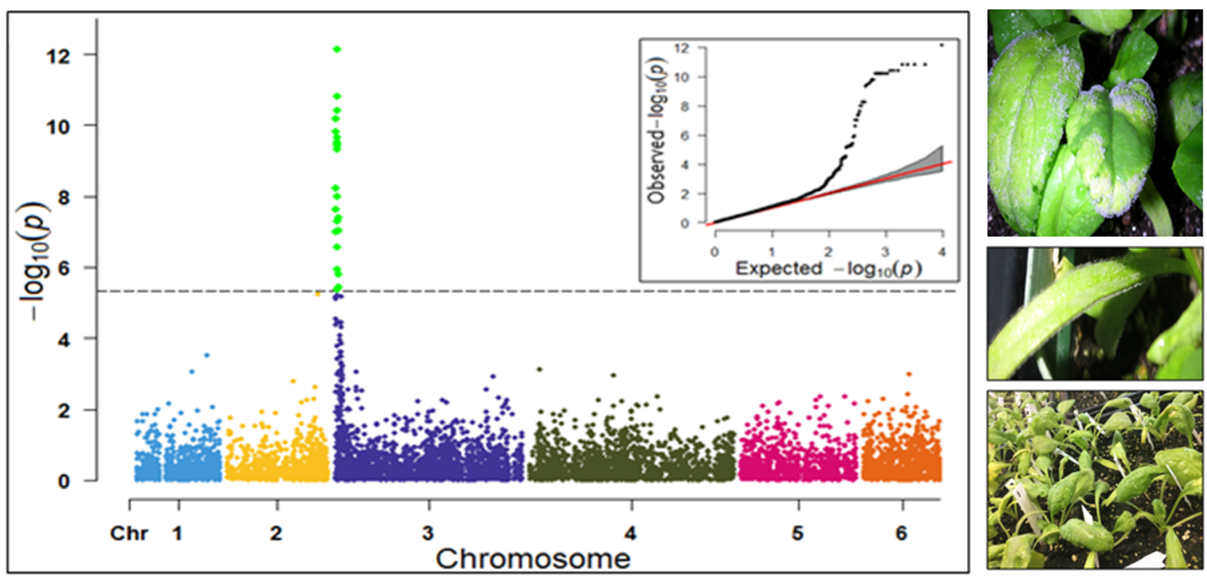 Figure 1. Manhattan and QQ plots showing significantly associated SNP markers on chromosome 3 to downy mildew race Pfs 13 resistance. Progenies from a cross of Lazio (Resistant) and Viroflay (Susceptible) were inoculated with Pfs race 5 to determine disease response. Association analysis performed with low coverage whole genome resequencing-generated SNP markers mapped the RPF2 locus between 0.47 to 1.46 Mb of chromosome 3 with peak SNP (Chr3_1221009) showing a LOD value of 61.6 in the TASSEL GLM model, which was within 1.08 Kb from gene Spo12821 that encodes CC-NBS-LRR disease resistance protein. The combined analysis of RPF2 and RPF3 segregating panels mapped the resistance region between 1.18-1.23 and 1.75-1.76 Mb of chromosome 3 (Figure 2) (Bhattarai et al. 2023). 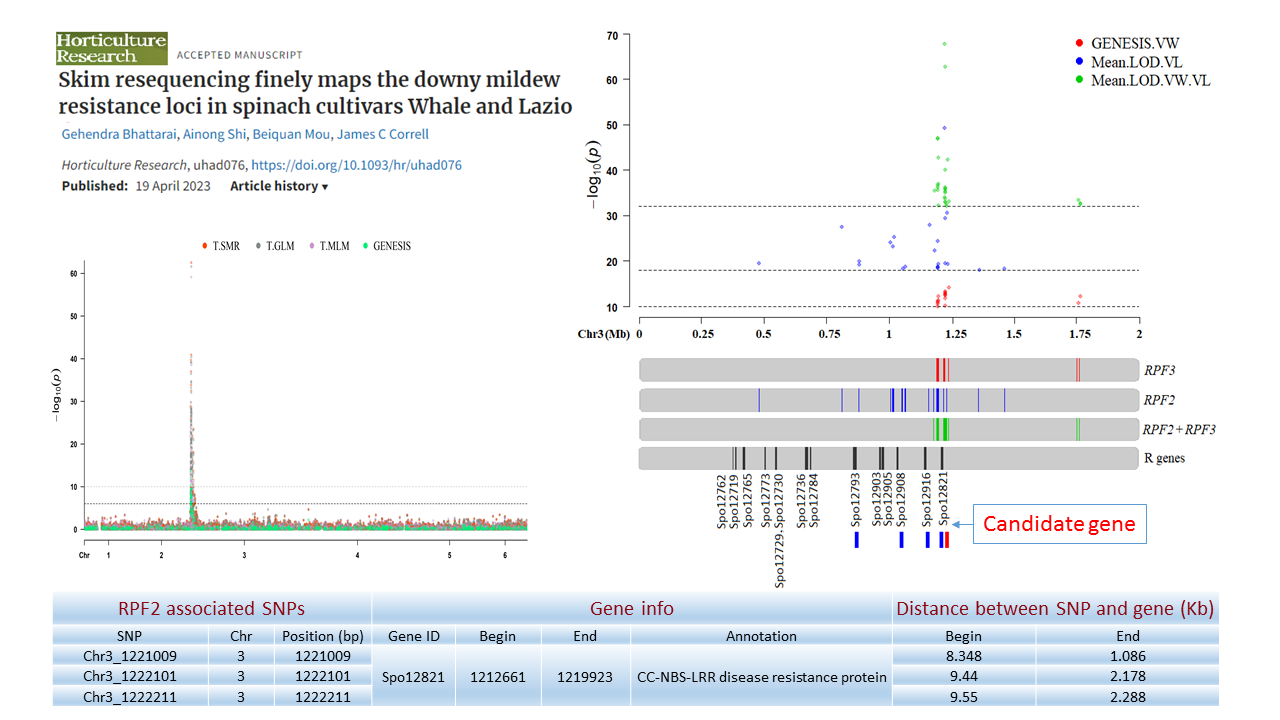 Figure 2. Manhattan plots showing significantly associated SNP markers and candidate genes identified on chromosome 3 to downy mildew race Pfs 5 resistance. Under field experiments, GWAS has been performed with >400 spinach genotypes comprising USDA germplasm accessions and commercial cultivars were evaluated for resistance to downy mildew pathogen between 2017–2019 in Salinas Valley, California and Yuma, Arizona. GWAS was performed using single nucleotide polymorphism (SNP) markers identified via whole genome resequencing (WGR) in GAPIT and TASSEL programs; detected 14, 12, 5, and 10 significantly associated SNP markers with the resistance from four tested environments, respectively; and the QTL alleles were detected at the previously reported region of chromosome 3 in three of the four experiments (Figure 3). In parallel, prediction accuracy (PA) was assessed using six genomic prediction (GP) models and seven unique marker datasets for field resistance to downy mildew pathogen across four tested environments. The results suggest the suitability of GS to improve field resistance to downy mildew pathogen (Bhattarai et al. 2022a). The RPF3 locus in the 1.22-1.23 Mb region of Sp75 chromosome 3 is 2.41-3.65 Kb from the gene Spo12821 annotated as NBS-LRR disease resistance protein (Bhattarai et al. 2022b). 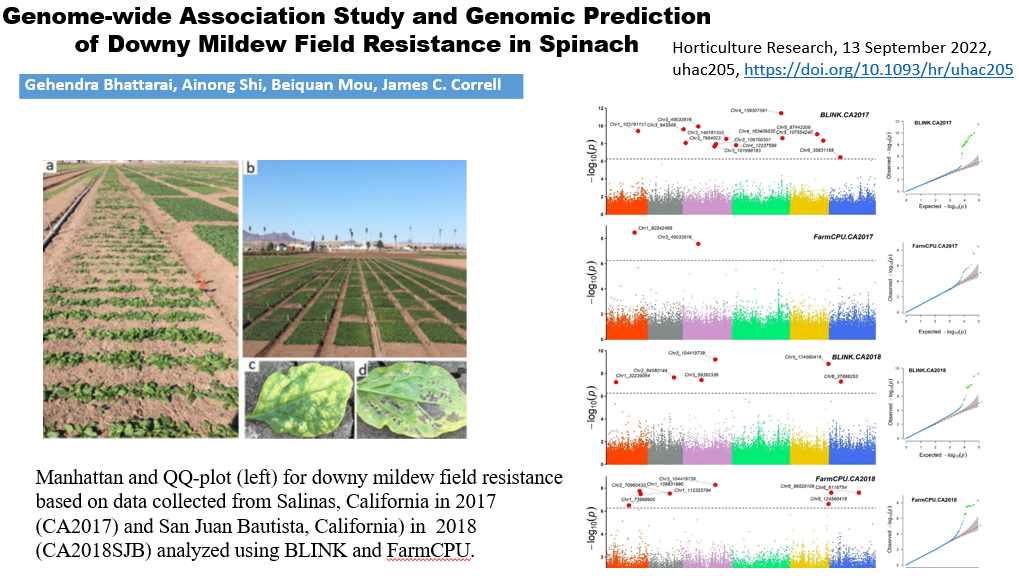 Figure 3. Field experiments for evaluating downy mildew resistance (left); QQ and Manhattan plots based on four GWAS models (right). In another experiment, GWAS performed under greenhouse/growth chamber conditions for resistance to Pfs 5 in 251 genotypes, including 216 USDA spinach germplasm accessions and 35 commercial hybrids/cultivars; the major QTL/alleles was mapped near the previously mapped region on chr. 3; 14 SNP markers were strongly associated with DM resistance; and the prediction accuracy (r-value) was 0.81 – 0.90 when 14 SNP markers were used (Olaoye 2021 MS Thesis; Olaoye et al. 2023) (Figure 4). 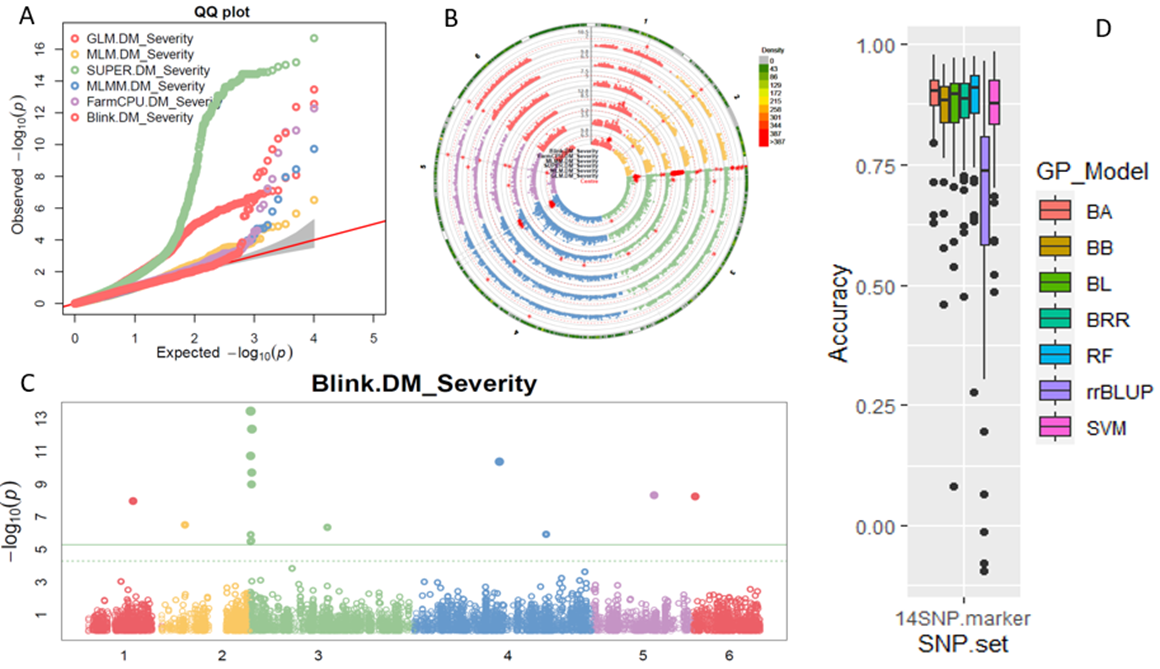 Figure 4. QQ and Manhattan plots based on six GWAS models (A, B); BLINK Manhattan plot (C) showing significantly associated SNP markers on chr 3 and other chr 1, 2, 4, 5, and 6 for downy mildew race Pfs 5 resistance; genomic prediction (r-value) of seven models based on 14 SNP markers. 2. White Rust Dr. Shi, Avila and Correll have performed GWAS for two panels for WR resistance. The first GWAS panel consisted of 464 spinach lines developed from four seed companies (Pop Vriend, Enza Zaden, Rijk Zwaan, and Sakata) and the University of Arkansas. The results showed that 107 lines were WR resistant (Figure 5 A); genetic diversity analysis showed that there were three clustered populations (Figure 5 B); nine SNPs were associated with WR resistance with a predictive accuracy (r) above 0.55. We have made presentations several times and the final report has been sent to companies (Shi et al. 2020, 2019, 2018a, b). 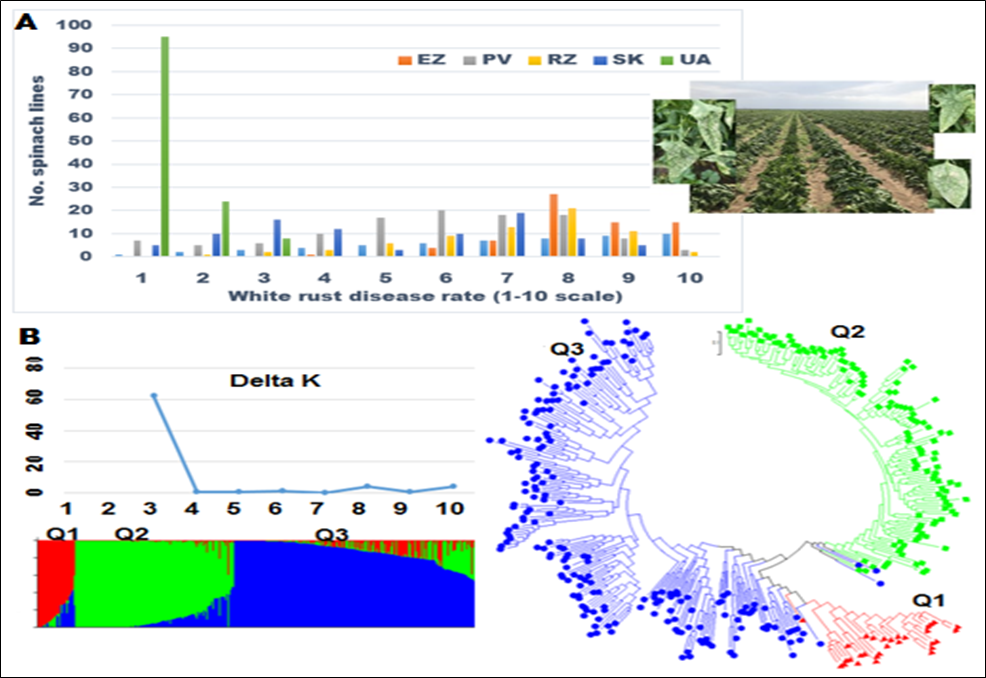 Figure 5. Screening results for WR resistance: (A) Distribution of disease severity index on evaluated germplasm and (B) Population structure. In the second GWAS panel consisting of 346 USDA germplasm accessions, 23 accessions were resistant; 40 and 9 SNPs were associated with WR resistance based on nine GWAS models, individually or combined; and predictive accuracy was up to 0.84 when 4,836 SNPs were used; 0.75 when the 40 SNP markers were used; and 0.61 when the 9 SNP markers were used based on nine GP models (Figure 6) (Shi et al. 2022). In addition, minor alleles were shown to be associated with WR susceptibility in the spinach study from PF Avila’s lab (Awika et al. 2019). 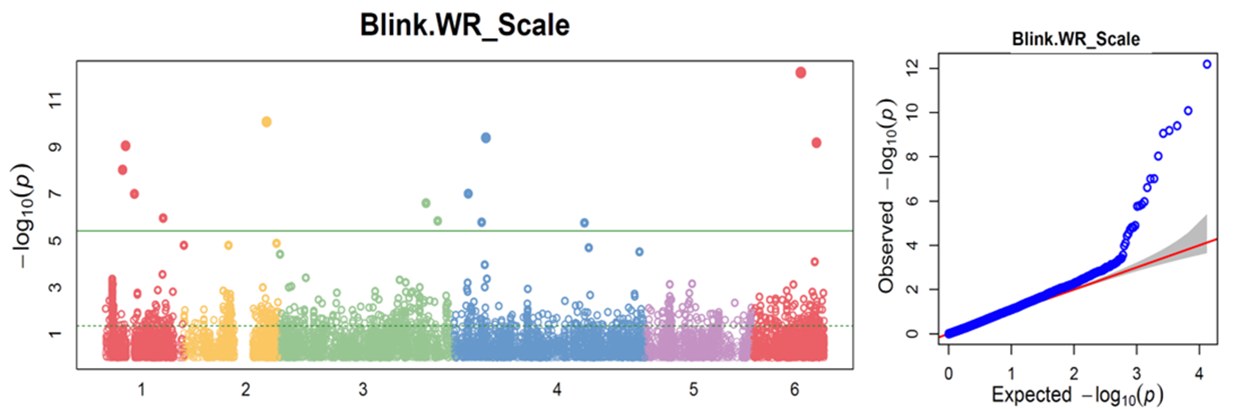 Figure 6. Manhattan and QQ plots showing significantly associated SNP markers on chromosomes 1, 2, 4, and 6 for white rust resistance. 3. Fusarium Wilt More than 350 USDA accessions and commercial cultivars, mainly comprising S. oleracea genotypes, were phenotyped by du Toit’s lab (Co-PI) (Gyawali et al. in preparation). GWAS was performed to identify a set of associated markers (Gyawali et al. 2019a, b, c) (Figure 7). In addition to the cultivated germplasm panel, 68 wild spinach (S. turkestanica) accessions plus 16 selected S. oleracea accessions were evaluated for resistance against three isolates of Fos (Fus058, Fus254, Fus322). The Fos-phenotyped panels, including wild accessions (Gyawali et al. 2021), were genotyped using GBS, and GWAS analysis identified SNP markers associated with multiple sources of excellent resistance to FW (Gyawali et al. in preparation). 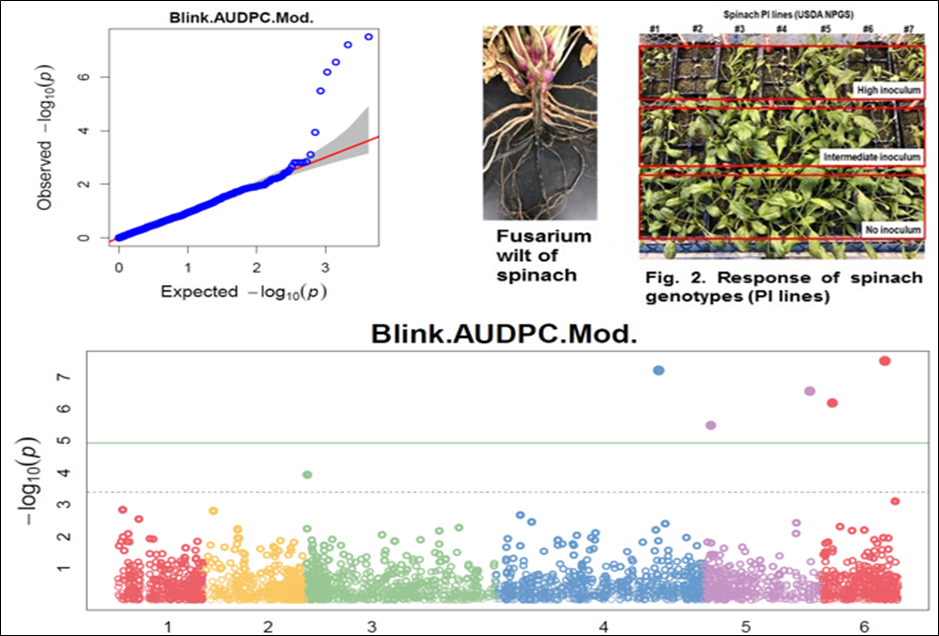 Figure 7. Manhattan and QQ plots showing significantly associated SNP markers on chromosome 4, 5, and 6 to Fusarium wilt resistance. 4. Stemphylium Leaf Spot A total of 271 USDA spinach germplasm accessions plus 35 commercial spinach cultivars and 5 University of Arkansas breeding lines, were evaluated for resistance to SLS using isolate Sb-1-St001 of S. vesicarium under greenhouse conditions. The results showed that 15 lines were resistant, and subsequent GWAS analysis using disease scores and whole WGR-generated SNP markers identified 42 SNP markers significantly associated with SLS resistance. One associated SNP marker was less than 7 Kb from gene SOV3g003360 annotated as NB-ARC domain leucine-rich repeat (LRR) with the putative functions known to impart disease resistance in plants. Six GS models evaluated across eight SNP datasets identified 42 GWAS-associated SNP sets implemented in the Bayesian ridge regression model as most promising to predict resistance to Stemphylium leaf spot with a prediction accuracy of 0.79 (Figure 8). Markers reported in this study will aid the development of Stemphylium resistant spinach cultivars through marker-assisted selection (MAS) and genomic selection (GS) (Liu et al. 2020, 2021; Bhattarai et al. 2022b). 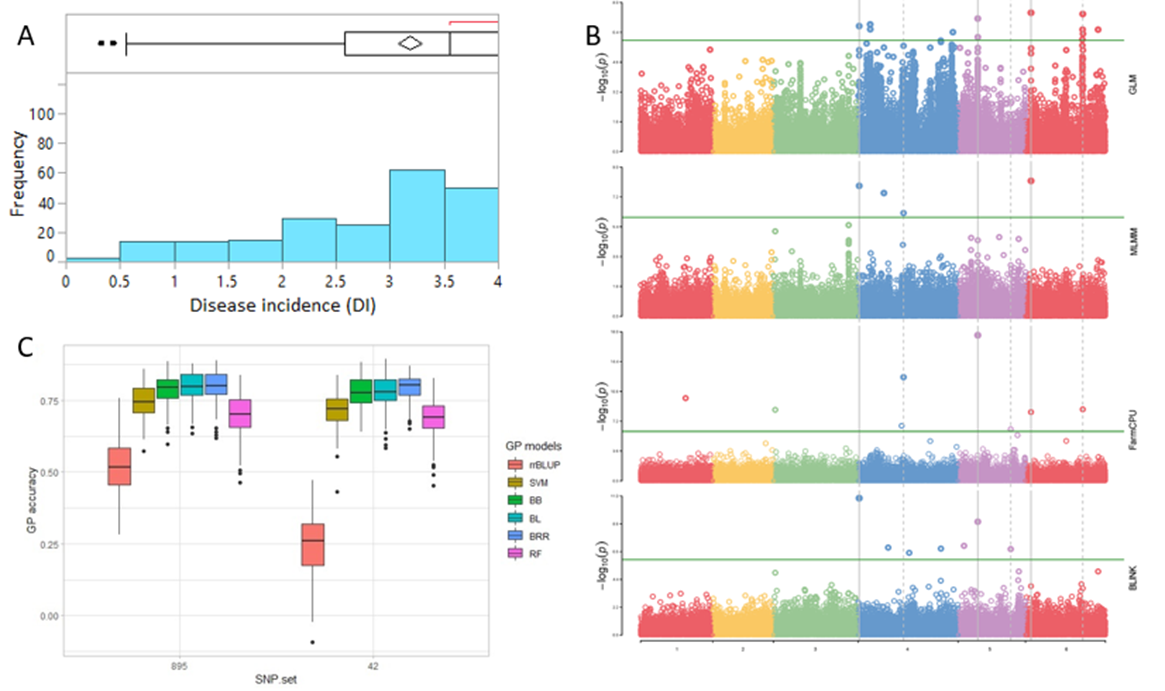 Figure 8. Distribution of Stemphylium leaf spot (SLS) disease incidence (A); Manhattan plots based on four GWAS models (Blink, FarmCPU. MLM, and GLM) showing significantly associated SNP markers on chr 4, 5, and 6 for SLS resistance (B); and genomic prediction (r-value) of six models based on 42 SNP markers (C). 5. Anthracnose Leaf Spot A diverse collection of 276 spinach accessions was scored for anthracnose disease severity. Alleles in linkage disequilibrium were tagged in haplotype blocks, and anthracnose-associated molecular markers were identified using single-SNP (sSNP), pairwise haplotype (htP) and multi-marker haplotype (htM) SNP tagging approaches. We identified 49 significantly associated markers distributed on several spinach chromosomes using all methods. The sSNP approach identified 13 markers, while htP identified 24 (~63% more) and htM 34 (~162% more). Of these markers, four were uniquely identified by the sSNP approach, nine by htP and nineteen by htM (Figure 9). The results indicate that resistance to anthracnose is polygenic and that haplotype-based analysis may have more power than sSNP. Using a combination of these methods can improve the identification of molecular markers for spinach breeding (Awika et al. 2020). 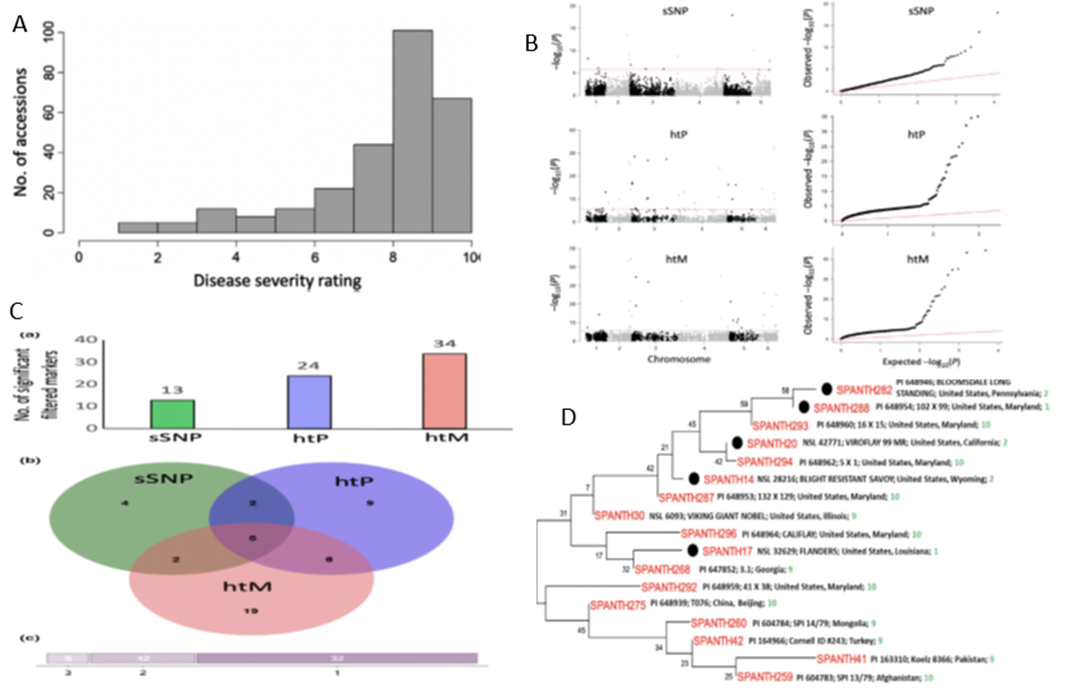 Figure 9. Distribution of anthracnose leaf spot (ALS) disease severity rating incidence (A); Manhattan and QQ-plots based on three GWAS analyses showing significantly associated SNP markers for ALS resistance (B); unique and overlapped significant markers associated with anthracnose resistance (C); and ancestry relationships of spinach accessions with respect to 49 significant polymorphic sites (SNP markers) (D). |
|
References (*corresponding author): 1. Awika, H, Cochran, V. Joshi, R. Bedre, K.K. Mandadi, C.A. Avila*. 2020. Single-marker and haplotype-based association analysis of anthracnose (Colletotrichum dematium) resistance in Spinach (Spinacia oleracea). Plant Breeding 139:402-418. https://doi.org/10.1111/pbr.12773 2. Awika, H.O., T.G. Marconi, R. Bedre, K.K. Mandadi, C.A. Avila*. 2019. Minor Alleles are Associated with White Rust Susceptibility in Spinach. Horticulture Research 6:129. https://doi.org/10.1038/s41438-019-0214-7 3. Bhattarai, G.*, A. Shi*, B. Mou*, and J. Correll*. 2023. Skim resequencing of progeny population of Lazio and Whale crossed with Viroflay finely mapped downy mildew locus and identified resistance genes in spinach Horticulture Research, uhad076, https://doi.org/10.1093/hr/uhad076 4. Bhattarai, G.*, A. Shi*, B. Mou*, and J. Correll*. 2022a. Resequencing worldwide spinach germplasm identifies downy mildew field tolerance QTLs and genomic prediction tools. Horticulture Research, Published: 13 September 2022, uhac205, https://doi.org/10.1093/hr/uhac205 5. Bhattarai, G., D. Olaoye, B. Mou*, J. C. Correll*, A. Shi*. 2022b. Mapping and selection of downy mildew resistance locus RPF3 in spinach by low coverage whole genome sequencing. Frontiers in Plant Science, PUBLISHED 06 October 2022, https://doi.org/10.3389/fpls.2022.1012923. 6. Bhattarai, G., W. Yang, A. Shi*, C. Feng, B. Dhillon, J.C. Correll*, and B. Mou*. 2021a. Mapping and candidate gene identification of downy mildew race 16 resistance in spinach. BMC Genomics 22-478. https://doi.org/10.1186/s12864-021-07788-8. 7. Bhattarai, G., A. Shi*, C. Feng, B. Dhillon, B. Mou*, J.C. Correll*. 2020a. Genome-wide association studies in multiple spinach breeding populations refine downy mildew race 13 resistance genes. Frontiers in Plant Science Volume 11 - 2020 https://doi.org/10.3389/fpls.2020.563187. 8. Liu, B., L. Stein, K. Cochran, L.J. du Toit, C. Feng, and J.C. Correll*. 2021. Three New Fungal Leaf Spot Diseases of Spinach in the United States and the Evaluation of Fungicide Efficacy for Disease Management. Plant Disease 105(2):316-323. https://doi.org/10.1094/pdis-04-20-0918-re 9. Liu, B., L. Stein, K. Cochran, L.J. du Toit, C. Feng, B. Dhillon, and J.C. Correll*. 2020. Characterization of Leaf Spot Pathogens from Several Spinach Production Areas in the United States. Plant Disease 104(7): 1994-2004. https://doi.org/10.1094/pdis-11-19-2450-re. 10. Olaoye, D. 2021. Resistance Screening and Association Analysis of Downy Mildew Resistance in Spinach (MS Thesis at Shi’s and Correll’s Lab), University of Arkansas. 11. Shi, A.*, G. Bhattarai, H. Xiong, A. Carlos*, C. Feng, B. Liu, V. J. Joshi, L. Stein*, B. Mou*, L.J. du Toit*, J.C. Correll*. 2022. Genome-wide association study and genomic prediction of white rust resistance in USDA GRIN Spinach Germplasm. Horticulture Research, Volume 9, 2022, uhac069, https://doi.org/10.1093/hr/uhac069. Submission/Preparation 12. Bhattarai, G., Liu, B., Shi, A.*, Correll, J.C*. 2022b. Genome-wide association analysis and genomic selection of Stemphylium vesicarium leaf spot resistance in USDA spinach germplasm. Preparing to submit in Frontiers in Plant Science, in an internal review. 13. Gyawali, S., … L.J. du Toit*, et al. Identification of major QTLs associated with Fusarium wilt resistance in wild spinach, Spinacia turkestanica (in preparation). 14. Olaoye, D., A. Shi*, J.C. Correll* et al. 2023. Genome-wide association study and genomic prediction of resistance to downy mildew race 15 in spinach (in preparation for Frontiers in Plant Science). Abstract/Presentation 15. Gyawali, S., G. Bhattarai, A. Shi, C. Kik, and L.J. du Toit. 2021c. Genetic Diversity and population Structure of Spinacia turkestanica, a Wild Progenitor of Cultivated Spinach, Spinacia oleracea. 2021 ASHS International Conference, August 5-9, Denver, Colorado. https://ashs.confex.com/ashs/2021/meetingapp.cgi/Paper/35994. 16. Gyawali, S., L. du Toit, J.C. Correll, and A. Shi. 2019a. Genome wide association studies of Fusarium wilt resistance in spinach (Spinacia oleracea L.). Poster presented at the American Phytopathological Society (APS) Annual Meeting, Plant Health 2019, 3-7 August 2019, Cleveland, OH. Abstract. https://apsnet.confex.com/apsnet/2019/meetingapp.cgi/Paper/13443. 17. Gyawali, S., and L. du Toit. 2019b. Spinach Fusarium wilt resistance assessment and mapping of QTL associated with wilt resistance. Western Washington Seed Workshop held at the Washington State University (WSU) Mount Vernon Northwestern Washington Research and Extension Center (NWREC) on 11 January 2019. 18. Gyawali, S., and L. du Toit. 2019c. Progress of Fusarium wilt components of this spinach SCRI project in the WSU Mount Vernon NWREC Field Day on 11 July 2019. 19. Shi, A., J. Correll, G. Bhattarai, B. Liu, C. Feng, H. Awika, C.A. Avila, and B. Mou. 2020. Evaluation and Genome-wide Association Study for White Rust Resistance in USDA Spinach Germplasm. ASHS 2020 Annual Conference. August 9-13, 2020, Orlando, FL. 20. Shi, A., J. Correll, C. Feng, B. Mou, C.A. Avila, L.A. Stein, R. Hogan, L. du Toit, J. Qin, G. Bhattarai, H. Awika, S. Gyawali, and S. Kandel. 2019. Progress at Developing Genetic and Molecular Resources to Improve Spinach Production and Management. HortScience 54, S137. (Abstr.). ASHS 2019 Annual Conference. July 21-25, Las Vegas Nevada. https://ashs.confex.com/ashs/2019/meetingapp.cgi/Paper/29998. 21. Shi, A. 2018a. Genome-wide association study and genomic selection in spinach. Gust talk on February 23, 2018a at ABI, Arkansas State University, Jonesboro, AR. 22. Shi, A., J. Qin, Y. Weng, J. Correll, C. Feng, G. Bhattarai, W. Ravelombola, B. Zia, W. Zhou, and B. Mou. 2018b. Genetic diversity, genome-wide association study and genomic selection in spinach. The 2018 International Spinach Conference, 02/14-02/15, 2018 in Murcia, Spain. Genetic diversity, genome-wide association study and genomic selection in spinach |